Humanized SpCas9 mRNA for Precise Gene Editing | CRISPR Cas9 Variants Explained
Harnessing Precision: Humanized SpCas9
The CRISPR-Cas9 system, originally discovered as a bacterial adaptive immune mechanism, has been ingeniously adapted into a powerful genome editing platform capable of precisely modifying DNA across a wide range of organisms, including humans. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) acts as a molecular memory in bacterial genomes, enabling defense against viral invasions.
Compared to previous tools like Zinc Finger Nucleases (ZFNs) and TALENs, CRISPR-Cas9 is more cost-effective, easier to use, and highly efficient. Its simplicity has democratized genome editing and catalyzed innovation in biomedical research, disease modeling, and therapeutic development. Notably, the first CRISPR-based therapy, Casgevy, has already gained regulatory approval for treating sickle cell disease.
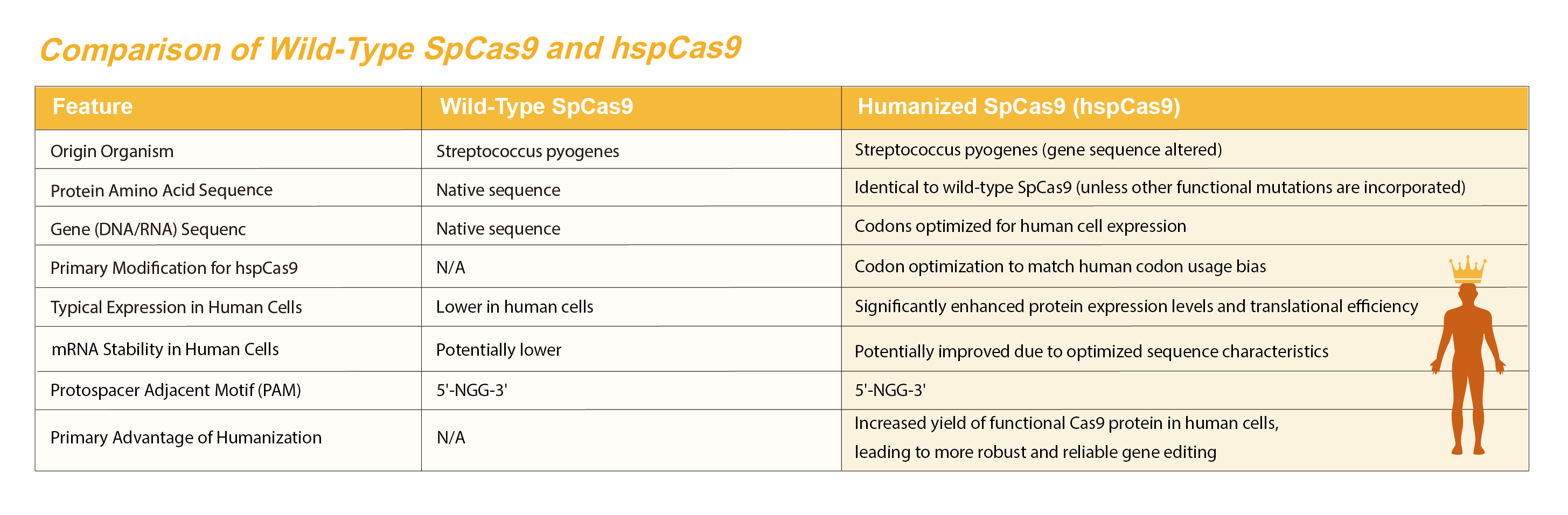
CRISPR-Cas9 gene editing has revolutionized modern molecular biology by enabling precise, efficient, and programmable modification of genomic sequences. Among the various CRISPR-associated nucleases, Streptococcus pyogenes Cas9 (SpCas9) remains the most widely used. To improve expression and reduce immunogenicity in human cells, a codon-optimized, humanized version—hSpCas9—was developed. This enhanced version facilitates high-level expression and efficient gene editing in mammalian systems, making it ideal for both research and therapeutic applications.

- The Prototypical Nuclease Streptococcus pyogenes Cas9 (SpCas9)
- The Need for Optimization: Introducing Humanized SpCas9 (hspCas9)
This "humanization" primarily refers to codon optimization for expression and can be a backbone for further protein engineering (e.g., high-fidelity variants). While hspCas9 improves expression , it doesn't inherently make the SpCas9 protein less immunogenic, as the amino acid sequence remains largely the same ; immunogenicity is a separate challenge addressed by protein engineering or delivery methods.
- The molecular mechanism of SpCas9
- Step-by-Step Mechanism of Action
.png)
The molecular structure of hSpCas9 has been optimized to function efficiently in human cellular environments. The protein consists of multiple domains that work together to achieve precise DNA cleavage, including the RuvC and HNH nuclease domains responsible for cutting the DNA strands. The humanization process involves modifications to improve protein folding, stability, and expression in mammalian cells. Advanced formulations of hSpCas9, such as those produced through in vitro transcription with Cap1 modifications and poly(A) tails, are designed to mimic natural mature mRNAs, enhancing both stability and translational efficiency.

Enhanced hspCas9 expression has made it vital for diverse biomedical applications.

Specific examples demonstrate the impact of hspCas9 and similar optimized Cas9 systems.



